Current Research
Leprosy
T cells are required for protection against intracellular infections, such as leprosy and tuberculosis; however, the mechanisms by which T cells contribute to host immunity remain unresolved. Leprosy, caused by the intracellular pathogen Mycobacterium leprae (mLEP), represents an accessible model to investigate human immune responses to intracellular bacterial infection. The spectrum of clinical manifestations of leprosy correlates with the immune response to mLEP. Patients with tuberculoid leprosy (T-lep) develop protective immunity and eliminate the infection, whereas those with lepromatous leprosy (L-lep) sustain a progressive infection. We established that T-cell subsets differed dramatically between blood and lesions, indicating the importance of studying the immune response at the site of disease. We seek to characterize antimicrobial T cells and their roles in reducing the viability of intracellular pathogen.
Potential projects:
- Elucidate mechanisms by which CD8+ cytolytic T lymphocytes (CTL) are triggered via TCR-dependent and/or -independent antimicrobial response
- Define the heterogeneity within the Th17 compartment of patients with T-lep and L-lep to map the IL-1β-mediated antimicrobial response
- Identify the innate pathways that induce antimicrobial T-cell differentiation in leprosy.
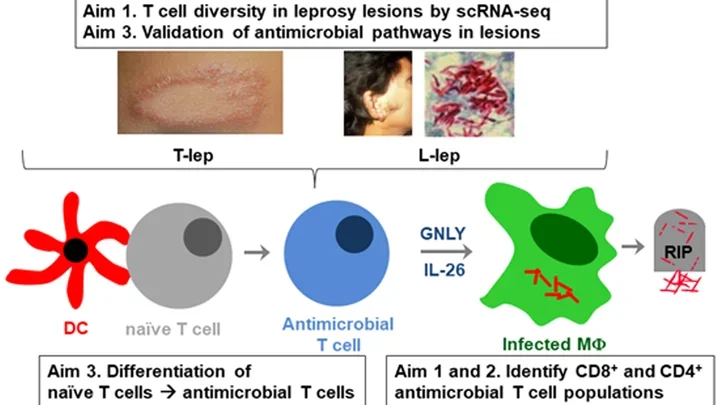
Leprosy: Schematic Details
The schematic diagram illustrating a study on the diversity and function of T cells in leprosy lesions focuses on three main aims:
- T cell diversity in leprosy lesions by single-cell RNA sequencing (scRNA-seq)
- Identification of CD8+ and CD4+ antimicrobial T cell populations
- Validation of antimicrobial pathways in lesions and differentiation of naïve T cells into antimicrobial T cells
The top row depicts:
- Images of leprosy lesions: Two photographs, one showing a leprosy lesion on the skin and another showing an individual with a leprosy lesion on the face. A third image shows the bacteria Mycobacterium leprae, the causative agent of leprosy.
- Labels "T-lep" and "L-lep": Likely referring to different types of leprosy or different stages of the disease.
The middle row depicts:
- Dendritic Cell (DC): Illustrated as a red dendritic cell interacting with a gray naïve T cell.
- Naïve T Cell: Shown transitioning into an antimicrobial T cell.
- Antimicrobial T Cell: Blue cell that is part of the immune response.
- Infected Macrophage (MΦ): A green cell infected by Mycobacterium leprae.
- GNLY and IL-26: Molecules (Granulysin and Interleukin-26) released by the antimicrobial T cell that help in killing the bacteria.
- RIP: Symbol indicating the death of the bacteria within the infected macrophage.
The bottom row depicts:
- Aim 3: Differentiation of naïve T cells into antimicrobial T cells.
- Aim 1 and 2: Identification of CD8+ and CD4+ antimicrobial T cell populations.
Acne: A Disease of Lipid Metabolism, Microbiome, and the Immune Response
Using acne as a model to study the interaction between the microbiome and the local immune response, we aim to investigate how crosstalk among molecular gene programs, distinct phylotypes of commensal bacteria Cutibacterium acnes (C. acnes), and surrounding cells in the microenvironment promote inflammatory innate and adaptive immune response.
Potential projects:
- Determine how metabolites and molecular signals released during inflammation affect the acne cellular microenvironment
- Investigate the mechanisms of distinct C. acnes phylotypes to promote inflammation or maintain homeostasis
- Identify specific genetic elements of commensal C. acnes that are responsible for the differential induction of cytokine and immune signaling
- Defining the influence of lipid metabolism in immune cell fate and function
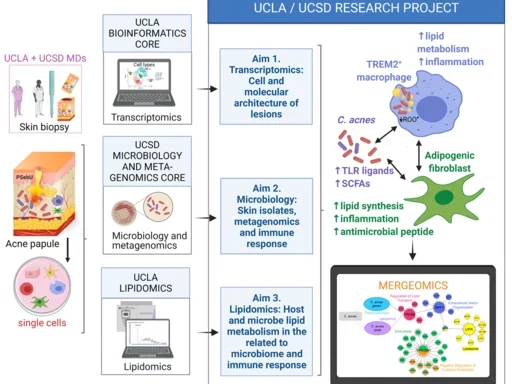
Acne: Schematic Details
Left Column
- UCLA + UCSD MDs:
- Skin biopsy: Visual representation of medical doctors taking a skin biopsy from patients.
- Acne papule: Illustration of an acne lesion.
- Single cells: Depiction of isolated single cells from the biopsy.
- UCLA Bioinformatics Core:
- Transcriptomics: Image of a computer with data analysis, indicating the use of transcriptomics to study gene expression.
- UCSD Microbiology and Meta-genomics Core:
- Microbiology and metagenomics: Visual of petri dishes and microbial cultures, highlighting the analysis of skin isolates and microbial communities.
- UCLA Lipidomics:
- Lipidomics: Illustration of a mass spectrometer, indicating lipidomic analysis to study lipid profiles.
Middle Column (Research Aims):
- Aim 1: Transcriptomics:
- Cell and molecular architecture of lesions: Study of gene expression in skin lesions.
- Aim 2: Microbiology:
- Skin isolates, metagenomics, and immune response: Investigation of the skin microbiome, including bacterial isolates, and how they influence the immune response.
- Aim 3: Lipidomics:
- Host and microbe lipid metabolism related to microbiome and immune response: Analysis of lipid metabolism in host and microbes to understand its impact on the microbiome and immune system.
Right Column (Mechanistic Insights):
- TREM2+ Macrophage:
- Increased lipid metabolism and inflammation: Illustration of a macrophage with TREM2 receptor, showing its role in lipid metabolism and inflammation.
- C. acnes (Cutibacterium acnes):
- TLR ligands and SCFAs: Depiction of C. acnes bacteria releasing toll-like receptor ligands and short-chain fatty acids (SCFAs), contributing to inflammation and immune response.
- Adipogenic Fibroblast:
- Lipid synthesis, inflammation, antimicrobial peptide: Diagram showing how these fibroblasts contribute to lipid synthesis, inflammation, and production of antimicrobial peptides.
- Mergeomics:
- Integration of data: Visual of data integration from transcriptomics, metagenomics, and lipidomics to identify key pathways and interactions.
Pulmonary TB Granuloma
The emergence of multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis strains is an emerging global public health crisis that underscores the urgent need to understand immune mechanisms in human tuberculosis (TB), with the ultimate goal of developing new strategies for its prevention and treatment. Our study focuses on investigating TB granuloma heterogeneity and its role in disease outcome, as well as the potential for identifying biomarkers that can reliably predict response to chemotherapy.
Potential projects:
- Determine the cell subpopulations and molecular architecture of TB granuloma and correlate the presence of cellular subtypes with response of chemotherapy in responder and resistant patients with TB
- Identify the role of macrophage subpopulations in TB granulomas that promote antimicrobial responses against M. tuberculosis and the mechanisms by which macrophage kill intracellular M. tuberculosis
- Determine the contribution of T cell subpopulation to regulate surrounding macrophages repolarization and the effects of M. tuberculosis antimicrobial activity
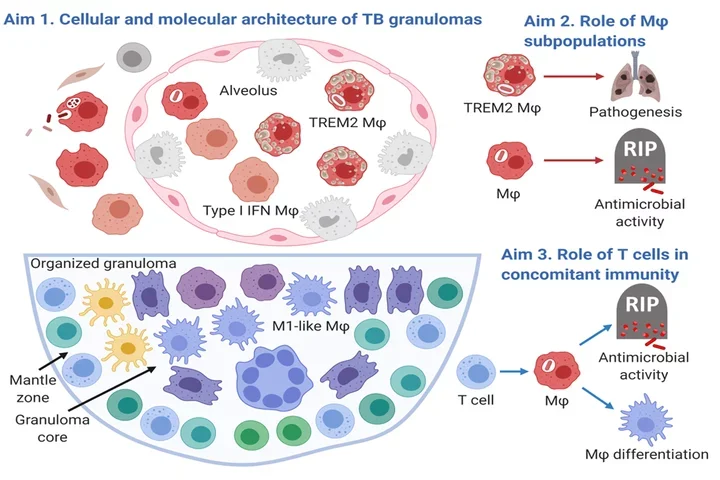
Pulmonary TB Granuloma: Schematic Details
Aim 1: Cellular and Molecular Architecture of TB Granulomas
- Upper Section:
- Alveolus: Shows a cross-section of an alveolus containing various types of cells.
- TREM2 Macrophages (MΦ): Red cells labeled as TREM2 MΦ, indicating a specific subtype of macrophages.
- Type I IFN Macrophages: Gray cells indicating another subtype of macrophages associated with Type I interferon responses.
- Other Cells: Various other cells within the alveolus, possibly representing different stages of TB infection and granuloma formation.
Aim 2: Role of Macrophage (MΦ) Subpopulations
- TREM2 MΦ and Pathogenesis:
- Shows TREM2 macrophages contributing to pathogenesis (disease progression), illustrated by an arrow pointing to a diagram of diseased lungs.
- Macrophage Antimicrobial Activity:
- Indicates the role of macrophages in killing the TB bacteria (RIP), with an arrow pointing to a macrophage engulfing and destroying the bacteria.
Aim 3: Role of T Cells in Concomitant Immunity
- Organized Granuloma:
- Illustration of a granuloma with a structured core and mantle zone.
- M1-like Macrophages: Dark purple cells labeled as M1-like MΦ, indicating a subtype involved in a pro-inflammatory response.
- T Cells: Various T cells surrounding the granuloma core, indicating their role in the immune response.
- T Cell Functions:
- Antimicrobial Activity: T cells contribute to the destruction of TB bacteria (RIP).
- Macrophage Differentiation: T cells influence the differentiation of macrophages, helping to form an effective immune response.
Overall Structure
- Granuloma Core: Dense central area of the granuloma with various immune cells.
- Mantle Zone: Outer area of the granuloma with different immune cells, including T cells and M1-like macrophages.
IL-26
Th17 cells defend the host against extracellular bacteria by releasing cytokines that recruit and activate a variety of other cells. Th17 cells mediate host defense against extracellular bacteria by releasing the antimicrobial protein interleukin (IL)-26 that kills bacteria in axenic cultures. Previous studies in our laboratory indicated that IL-26 expression in leprosy lesions significantly correlates with elimination of bacteria over time. In addition, we showed that IL-26 enters Mycobacterium leprae-infected macrophages, induces autophagy and phagolysosome fusion, colocalizes with the intracellular bacteria, and reduces bacterial viability. Our study aims to identify how IL-26 released by the activation of Th17 cells contribute to host defense intracellular bacteria such as bacterium M. leprae.
Potential projects:
- Determine how the extracellular IL-26 gain access to intracellular pathogens within macrophages and trigger an antimicrobial response
- Determine how IL-1b-activated secretion of IL-26 by Th17 cells contributes to the antimicrobial response against intracellular M. leprae and identify additional producers of IL-26
- Asses the role of IL-26 in host defense against various intracellular bacteria and whether distinct subcellular compartments are required for IL-26 antimicrobial activity
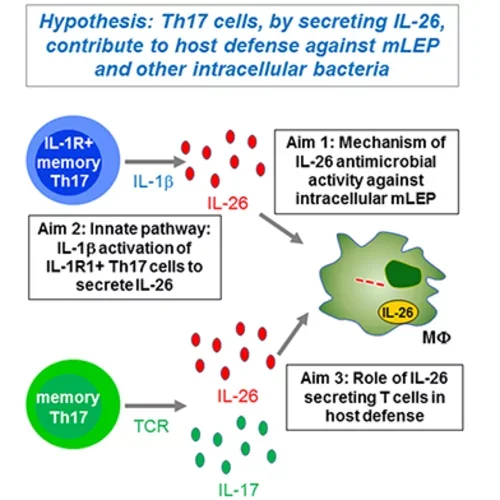
IL-26: Schematic Details
Hypothesis
Th17 cells, by secreting IL-26, contribute to host defense against mLEP and other intracellular bacteria.
Visual Components and Aims
- IL-1R+ memory Th17 cells
- These cells produce IL-1β.
- IL-1β
- This cytokine stimulates the secretion of IL-26 from IL-1R1+ Th17 cells.
- IL-26 Secretion
- IL-26 is depicted as red dots, representing its release from Th17 cells.
- Memory Th17 cells
- These cells can be activated via their T cell receptors (TCR) to secrete IL-17 (depicted as green dots) and IL-26.
Research Aims
- Aim 1: Investigate the mechanism of IL-26's antimicrobial activity against intracellular mLEP.
- Aim 2: Study the innate pathway involving IL-1β activation of IL-1R1+ Th17 cells to secrete IL-26.
- Aim 3: Explore the role of IL-26 secreting T cells in host defense.
Central Interaction
- The released IL-26 acts on macrophages (represented by MΦ), which is a key focus of the study.
CD1-Restricted T Cell Responses in Skin
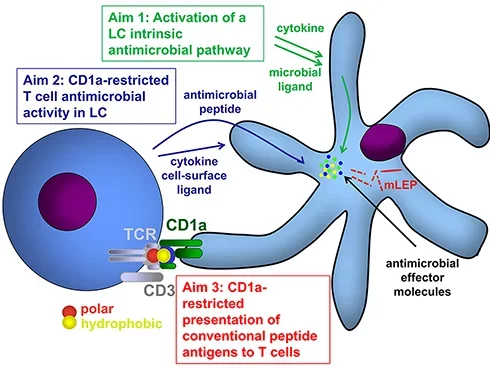
One crucial barrier mechanism via which the skin defends against microbial infection involves Langerhans cells (LCs), which are a skin-specific resident dendritic cell (DC) subset in the epidermis. Previous studies in our laboratory found that LCs contribute to the cutaneous immune response to Mycobacterium leprae, the causative agent of leprosy, via a langerin-dependent, CD1a-restricted antigen presentation pathway to T cells. We uncovered new paradigms of CD1 immunobiology, including defining the lipid-containing CD1-restricted T cell antigen structure and mechanisms of CD1 antigen presentation, mechanisms via which CD1 expression is regulated in skin, and the first evidence for direct T cell-mediated antimicrobial activity. To build on these findings, we aim to examine the role of CD1a-restricted T cell and LC in host defense in skin.
Potential projects:
- Determine specific pathways by which activated LCs kill intracellular bacteria as well as microbial ligands and their role in LC-mediated antimicrobial response
- Investigate mechanisms by which CD1a-restricted T cells kill intracellular pathogens in LCs
- Define mechanisms by which LCs process and present CD1a-restricted bacterial peptides to T cell
CD1: Schematic Details
Hypothesis
The hypothesis is centered around the functions of CD1a in immune responses, particularly in Langerhans cells (LC).
Visual Components and Aims
- Langerhans Cell (LC)
- The LC is depicted interacting with microbial ligands and cytokines (green arrows).
- Microbial Ligands and Cytokines
- These activate intrinsic antimicrobial pathways within the LC.
- CD1a Molecule
- CD1a is shown presenting antigens on the surface of the LC.
- CD1a binds to antigens, including both polar (red) and hydrophobic (yellow) components.
- Antimicrobial Peptides and Effector Molecules
- LCs release antimicrobial peptides (blue dots) and other effector molecules to combat mLEP.
- T Cell Interaction
- T cells interact with the LC via the T cell receptor (TCR) and CD3 complex, recognizing antigens presented by CD1a.
Research Aims
- Aim 1: Study the activation of an intrinsic antimicrobial pathway in LCs by microbial ligands and cytokines.
- Aim 2: Investigate the antimicrobial activity of CD1a-restricted T cells in LCs.
- Aim 3: Understand the CD1a-restricted presentation of conventional peptide antigens to T cells.
Central Interaction
- The interaction between the LC presenting antigens via CD1a and the T cell recognizing these antigens is crucial for the immune response against mLEP.